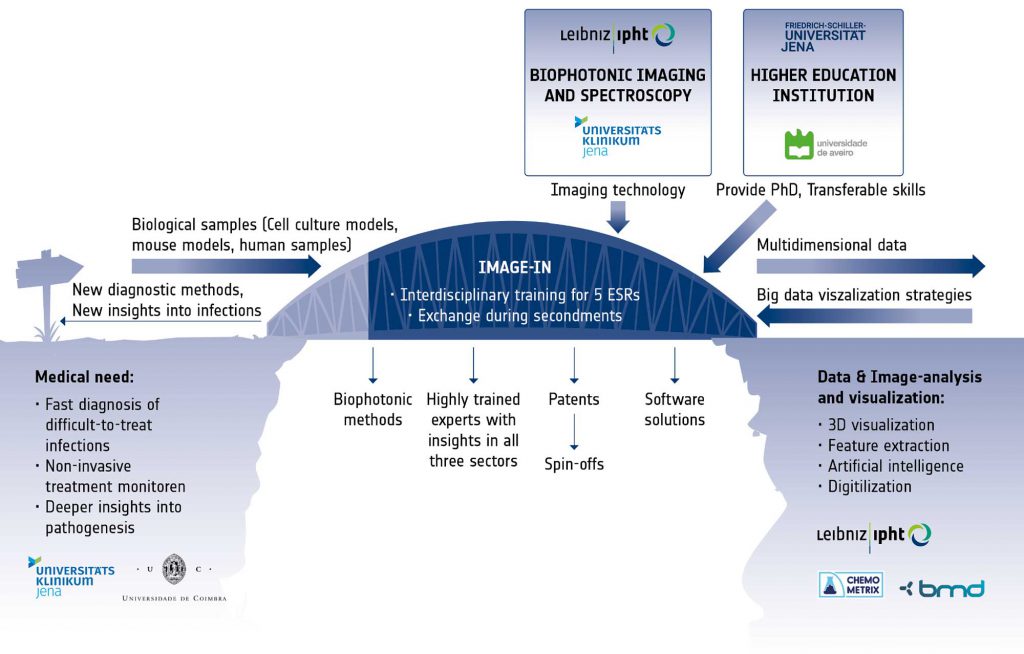
The successful realization of the complex objectives requires not only insights into the medical problem and into the physics of the different available imaging modalities, but also a deep understanding of handling large imaging data. Therefore, the IMAGE-IN training program will bridge the gap between the clinical view on the multiple facets of infection, the complex imaging technology and the statistical algorithms and software solutions, through the creation of a stable link between the disciplines and the assurance of vivid exchange between the involved sectors. With the IMAGE-IN program we want to spur interest and innovation to develop new imaging technologies for infectious disease and their translation to the clinic. The keystones of the IMAGE-IN research program are Excellence and Exchange. Five individual research projects are conducted by Early Stage Researchers (ESRs), supervised by three senior scientists from three different institutions covering the different sectors. Each trainee will benefit from individual coaching by his/her supervisors as well as a number of training workshops organised within the IMAGE-IN network. During their training, the ESRs will create their network in all disciplines, thus helping to overcome tardiness in technology transfer from the academic research to the industry for the successful medical application.
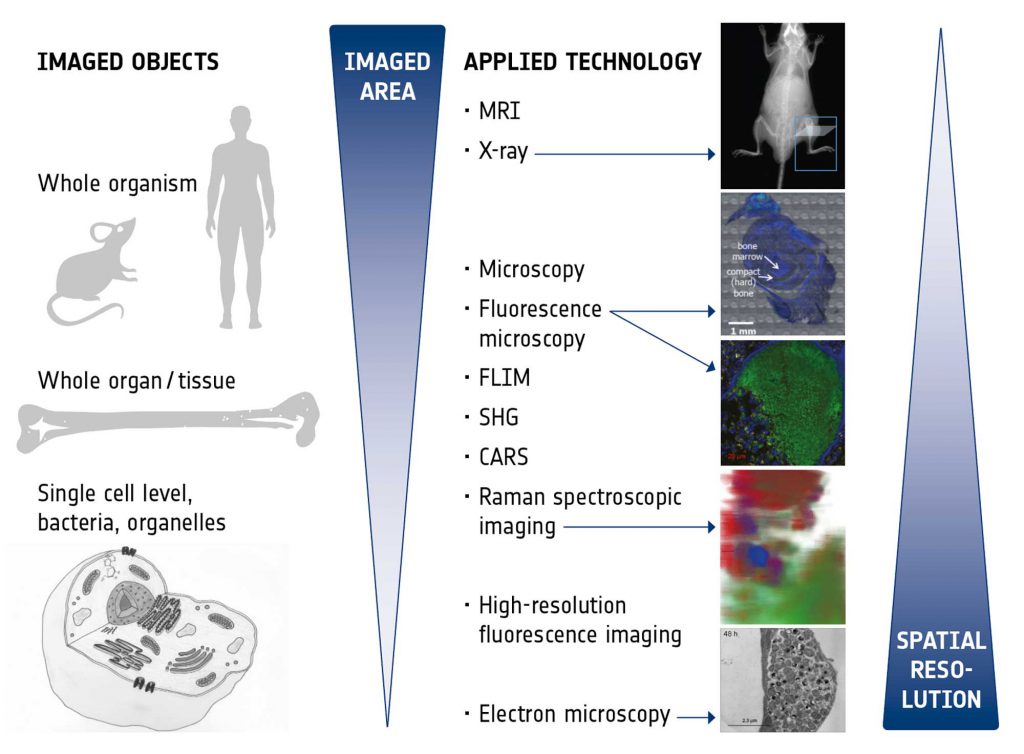
The imaging technologies to be exploited and correlated within this IMAGE-IN network were chosen to provide optimal insight into the niche of the pathogen within the host, reveal information on the host response, be suitable for future application as diagnostic tools to detect infections, and also to monitor treatment responses. Figure 2 presents an overview of the different technologies and their characteristics. The various imaging modalities are able to span different scales and all bring their unique advantages:
- Magnetic resonance imaging (MRI) provides excellent intrinsic soft tissue contrast, has unlimited penetration depth and, in addition, enables functional and metabolic assessment in vivo,
- X-ray imaging is a fast and easily applicable technique to assess hard tissue structures (bones) and the skeletal system in vivo and with high throughput,
- Microscopy with principle staining (e.g., H&E, Giemsa) is an established technology in clinical routine which enables fast and easy medical diagnostics like characterization of different tissues and pathological tissue transformations, classification of immune response and residing pathogens. In the time of emerging digital pathology it is undergoing significant transformation.
- Fluorescence microscopy enables live cell imaging of pathogens during infections in real time in cell culture models as well as the detection, identification and intracellular localization of pathogens in (fixed) tissue samples,
- Raman spectroscopic imaging provides insights into metabolic states of pathogens as well as changes in the surrounding host environment during infections by chemometric analysis in a label-free, non-destructive and space-resolved manner,
- Label-free, real-time, multimodal imaging (coherent anti-Stokes Raman (CARS), two photon excited fluorescence (TPF), fluorescence life time microscopy (FLIM), and second harmonic generation (SHG) enables (ultimately, intra-vital,) high resolution live cell imaging of metabolic states of pathogens, monitoring and controlling of the environmental changes, characterizing drug-cell interaction.
- High resolution fluorescence imaging provides details in sub-cellular localization of pathogens by specific labelling of pathogens and cellular compartments, enables the visualization of pathological changes in cellular structure and may also be used for high-resolution live-cell analysis of pathogens in their (changing) intracellular environment,
- Electron microscopy (here, in particular focussed ion beam scanning electron microscopy, FIB-SEM) yields highest resolution images to reveal the ultrastructure of host cells as well as of pathogens in their natural niche in the host (tissue, cell). Fixed tissue samples of infected animal models and cell culture models can be used.